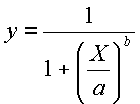| Parameter name | Description |
|---|
| Canopy Function Used | The probability distribution function to be used to distribute seeds in canopy conditions. For the behaviors Non-gap spatial disperse and Masting spatial disperse, these PDFs are always the ones used. |
| Lognormal Canopy X0 | The mean of the lognormal function under canopy conditions, or under non-masting conditions in the case of Masting spatial disperse (see equation below). This is only required if the canopy probability distribution function is lognormal. |
| Lognormal Canopy Xb | The standard deviation of the lognormal function under canopy conditions, or under non-masting conditions in the case of Masting spatial disperse (see equation below). This is only required if the canopy probability distribution function is lognormal. |
| Masting Disperse - Masting Beta | The β value under masting conditions. |
| Masting Disperse - Masting CDF "a" | The "a" value in the cumulative density function that is used to decide when masting events occur. |
| Masting Disperse - Masting CDF "b" | The "b" value in the cumulative density function that is used to decide when masting events occur. |
| Masting Disperse - Masting Group | Species in the same group always mast together. If all the group numbers are different, then each species masts separately. The actual numbers do not matter, just whether species have identical numbers. |
| Masting Disperse - Masting Lognormal X0 | The mean of the lognormal function under masting conditions. This is only required for a species if the canopy probability distribution function for that species is lognormal. |
| Masting Disperse - Masting Lognormal Xb | The standard deviation of the lognormal function under masting conditions. This is only required for a species if the canopy probability distribution function for that species is lognormal. |
| Masting Disperse - Masting STR Mean | The mean annual STR value under masting conditions. If the Masting Disperse - STR Draw PDF is Deterministic, then this is the STR value used. |
| Masting Disperse - Masting STR Standard Deviation | The standard deviation of the STR value under masting conditions. If the Masting Disperse - STR Draw PDF is Deterministic, then this value is not used. |
| Masting Disperse - Mast Proportion Participating (0-1) | The proportion of all adults for a species that participate in disperse during a masting timestep, as a value between 0 and 1. |
| Masting Disperse - Non-Masting Beta | The β value under non-masting conditions. |
| Masting Disperse - Non-Masting STR Mean | The mean annual STR value under non-masting conditions. If the Masting Disperse - STR Draw PDF is Deterministic, then this is the STR value used. |
| Masting Disperse - Non-Masting STR Standard Deviation | The standard deviation of the STR value under non-masting conditions. If the Masting Disperse - STR Draw PDF is Deterministic, then this value is not used. |
| Masting Disperse - Non-Mast Proportion Participating (0-1) | The proportion of all adults for a species that participate in disperse during a non-masting timestep, as a value between 0 and 1. |
| Masting Disperse - Masting Weibull Dispersal | The dispersal value for the weibull function under masting conditions. This is only required for a species if the canopy probability distribution function for that species is weibull. |
| Masting Disperse - Masting Weibull Theta | The θ for the weibull function under masting conditions. This is only required for a species if the canopy probability distribution function for that species is weibull. |
| Masting Disperse - Stochastic STR Draw Frequency | If the STR value is stochastic, this determines whether a new value is generated once per species per timestep or once per tree per timestep. If the Masting Disperse - STR Draw PDF is Deterministic, then this value is not used. |
| Masting Disperse - STR Draw PDF | Whether the STR value should be deterministic, or generated each timestep using a normal or lognormal distribution. |
| Minimum DBH for Reproduction, in cm | The minimum DBH at which a tree can reproduce. This value does not have to match the Minimum adult DBH. |
| Seed Distribution | The distribution method to be applied to seeds (randomization). The forms for these functions can be found here. Choices are:- Deterministic - no randomization.
- Poisson - use the number of seeds as the mean in a Poisson probability distribution function.
- Normal - use the number of seeds as the mean in a normal probability distribution function. You must then supply a standard deviation for the function.
- Lognormal - use the number of seeds as the mean in a lognormal probability distribution function. You must then supply a standard deviation for the function.
- Negative binomial - use the number of seeds as the mean in a negative binomial probability distribution function. You must then supply a clumping parameter.
|
| Seed Dist. Clumping Parameter (Neg. Binomial) | If you have chosen the negative binomial probability distribution function for "Seed distribution", this is the clumping parameter of the function, in seeds per m2. If you have not chosen that PDFs, then this parameter is not required. |
| Seed Dist. Std. Deviation (Normal or Lognormal) | If you have chosen the normal or lognormal probability distribution functions for "Seed distribution", this is the standard deviation of the function, in seeds per m2. If you have not chosen these PDFs, then this parameter is not required. |
| Weibull Canopy Dispersal | The dispersal value for the Weibull function under canopy conditions, or under non-masting conditions in the case of Masting spatial disperse (see equation below). This is only required if the canopy probability distribution function is Weibull. |
| Weibull Canopy Theta | The θ for the Weibull function under canopy conditions, or under non-masting conditions in the case of Masting spatial disperse (see equation below). This is only required if the canopy probability distribution function is Weibull. |
When the run starts, it is assumed a masting last event took place in timestep -1. A random number is used to determine whether a mast occurs in the current timestep. Disperse happens the same way in mast and non-mast timesteps, but the parameters used are different.
Species may be organized into groups to create synchrony in masting. The Masting Disperse - Masting Group parameter allows you to assign group numbers to species. The actual value of the group number is not important. It only matters if more than one species has the same number. If one species in a group masts, all species in that group do. Each group's mast decision is made separately, so sometimes more than one group may mast at a time. If all species have a different group number, then they all mast independently of one another.
If you choose to use a stochastic STR, the STR value can be generated once per species per timestep, or once per tree per timestep. If the value is generated once per species, all individuals of that species use the same STR value that timestep.
Once the behavior has decided whether masting occurs, and what the STR values are, then disperse proceeds exactly as described in the Non-gap spatial disperse behavior.
Apply this behavior to all trees of at least the minimum reproductive age for your chosen species. If the minimum reproductive age is less than the Minimum adult DBH, be sure to apply this behavior to saplings as well as adults.